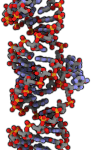
Kinetic scheme for force-induced dissociation of the 2-bp MMLV kissing-loop complex at 100 pN. (A) The initial conformation of the kissing loop based on the NMR structure; equal and opposite forces are applied to each strand. (B) Conformational rearrangement to the “bridging” form with H-bonds parallel to the applied force and stacked, flanking adenine residues (in red). (C) The top adenine “unstacks” and “restacks” an average of three times. (D) The top C–G base pair breaks and reforms an average of 12 times. (E) The bottom C–G base pair breaks and reforms an average of four times, rescued by the bottom adenine stack. (F) The final irreversible dissociation.
Mechanism of enhanced mechanical stability of a minimal RNA kissing complex elucidated by nonequilibrium molecular dynamics simulations AA Chen, AE García Proceedings of the National Academy of Sciences 109 (24), E1530-E1539
Summary:
Inspired by single-molecule optical tweezers experiments by Pan Li (UAlbany), we use molecular simulations with applied constant force to investigate the origin of the high mechanical stability of retroviral kissing-loop complexes. By mapping our the kinetic transitions between different force-induced intermediates, we are able to explain the unusual sequence specific stability of loop-loop complexes, in particular the requirement for unpaired, flanking purine bases which we find to be crucial for mechanical stability.